Abstract
Bayesian networks are graphical models that capture the conditional independences and dependences between random variables of a probability distribution. Specifically, each directed arrow specifies a direct conditional dependence between its two nodes.
Bayesian (or belief) networks represent conditional independence and dependence assertions over a complex joint distribution via a directed acyclic graph.
- Each node is a random variable that has conditional distribution
. We can think of this conditional as represented by probability tables. - Each directed edge is an conditional dependence assertion.
The following is an illustration of a simple network.
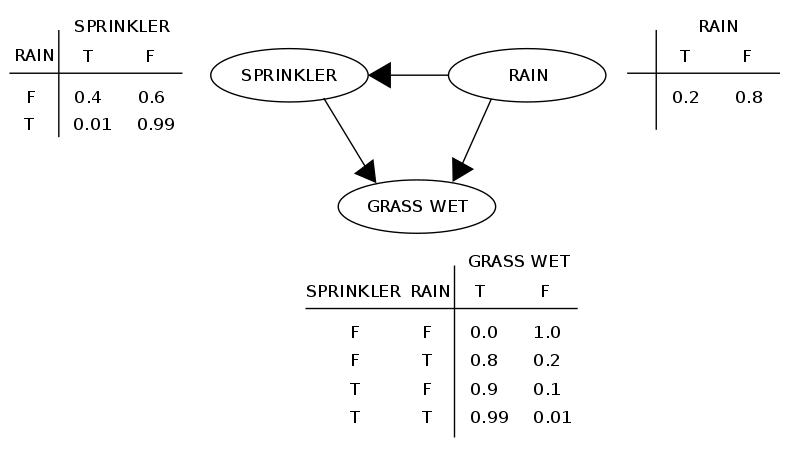
By representing conditional dependences using arrows, our graph captures both the conditional independences and dependences of the probability distribution. As a simple example, if we have a simple graph with two nodes
One variant, called ⏰ Dynamic Bayesian Networks, uses these variables to capture changes over time with the Markov assumption and time invariance.
Independencies
In more complex graphs, we can interpret the directed edges as follows.
- With
, we have but . The former is because knowing anything more about wouldn’t help us learn more about since affects but we know already. As for the latter, if we didn’t know , learning gives us information about and thus also about . - With
, we have but . This follows a similar reasoning as the previous example: knowing blocks the flow of information from to since our knowledge of would be redundant given , but not knowing allows the information to flow. - With
, we have but . The former is because knowing gives information about unknown , but more information about does not reveal anything about . On the other hand, if we did know , since both and affect , knowing would reveal some information about .
A common example for the third configuration is the alarm scenario:
Active Trails
Active trails represent flow of information; variables connected by an active trail are not conditionally independent. For each triplet
, not observed. , not observed. , not observed. , observed or one of its descendants is observed.
D-Separation
If there is no active trail between
Note
All d-separated independences in the graph are independencies in the probability distribution, but it’s not guaranteed that all independences in the distribution are represented in the graph.
I-Maps
Let
This is the case when
Note that we don’t require
Some graphs can also encode the same independences—we call them I-equivalent. By the independencies specified above, two graphs are I-equivalent if they have the same skeleton (same edges, not considering directionality) and same v-structures.
Info
A more connected network represents a distribution with few conditional independences. Each conditional independence limits the size of the class of distributions that satisfy it, so a more connected network models a larger class of joint probability distributions.
Ancestral Sampling
To sample from our distribution, we can go through the nodes from top to bottom. Specifically, we first topologically sort our variables, then go in order, sampling from each table. The sorting guarantees that we calculate all the conditionals in order.